 |

Citrus Canker In-Depth

I. Economic Hosts

II. Pathogens

III. Disease

IV. Symptoms and Signs:

V. Isolation:

VI. Identification:

Colonies of X. citri and X. campestris pv. aurantifolii have not been distinguished
on the basis of cultural characteristics from X. campestris pv. campestris strains.
Colonies on agar plates are circular, convex, semi-translucent and yellow, and the
margins are entire and standard determinative tests are used to identify strains of
the genus [13,14]. Crude methanol extracts (ten minutes at 65 C) of cells exhibit a
major absorption peak between 443 and 446 nm [7], which is diagnostic of the
xanthomonadin pigment and not found in other yellow bacteria. All strains of
X. citri form a highly clonal group and all strains of X. campestris pv.
aurantifolii form a different highly clonal group; the groups may be identified
and distinguished from all other xanthomonads by characteristic restriction
fragment length polymorphism (RFLP) profiles [7]. A detailed protocol on this
identification technique and its application to Xanthomonas has been published
[5]. The type strain of the species X. citri (3213, ATCC 49118) and pathotype
strains of X. campestris pv. aurantifolii (Pathotype B: XC69, ATCC 51301;
Pathotype C: XC340, IAPAR 9694, ICMP 8434, IBSBF 417, ATCC 51302) and X.
campestris pv. citrumelo (3048, ATCC 49120) are available through the American
Type Culture Collection for comparative purposes.

The use of RFLP data alone to formulate a taxonomy and reinstate X. citri to
species [7] has been criticized [21], but the reinstatement was not invalidated.
Most microbial taxonomists agree that phylogeny should determine taxonomy and
that "the phylogenetic definition of a species generally would include strains
with approximately 70% or greater DNA-DNA relatedness" [22]. Since X. citri
strains are only 30% similar to X. campestris 33913 (the type strain) [4], the
DNA-DNA hybridization data are consistent with the RFLP data, and the
reinstatement to species is consistent with the phylogenetic standard. The
taxonomic status of X. campestris pv. aurantifolii strains is unresolved. These
strains are only 37-40% related to X. campestris 33913, but are also only 62-63%
related to X. citri strains [4], form a distinct RFLP group [7], and differ
serologically from X. citri strains [1]. The expedient of lumping these relatively
distinct groups together as pathovars of X. citri may not be taxonomically sound.

Identical symptoms induced by two taxonomically distinct groups of strains is
indicative of a common pathogenicity factor. Gene pthA is essential for X.
citri to elicit cankers on citrus, and pthA confers this ability to various X.
campestris strains (for example, pathovars alfalfae and citrumelo) [19,20].
Functionally homologous genes (pthB and pthC) have also been identified and
cloned from X. campestris pv. aurantifolii pathotype B and pathotype C,
respectively [Gabriel, unpublished data]. Both pthB and pthC are essential
for X. campestris pv. aurantifolii pathotypes B and C, respectively, to cause
cankers on citrus, and pthB and pthC confer this ability to various X.
campestris strains [Gabriel, unpublished data]. All three genes are therefore
functionally interchangeable, and these genes may have been transferred
horizontally on plasmids between X. citri and X. campestris pv. aurantifolii
strains. Apparently homologous genes are found in all canker-causing strains,
and have not been found in non-canker inducing strains isolated from citrus,
such as X. campestris pv. citrumelo. Therefore a single common gene appears to
be diagnostic (and responsible) for a Xanthomonas strain's ability to induce
cankers on citrus.

|

|
|

|

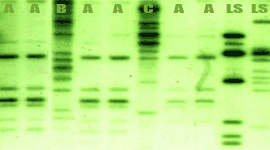
Blot Test
DNA probes, polymerase chain reaction (PCR)
primers and antibodies against the protein products of these genes may
provide rapid and certain diagnostic tests for xanthomonads capable of
causing cankers on citrus.
|

|
Genes pthA, pthB and pthC are all members of an avirulence / pathogenicity gene
family widely distributed in the genus Xanthomonas [3,20]. Avirulence genes
determine race specificity and can determine host range [8]. Genes pthA, pthB
and pthC, when transferred into X. citri, X. campestris pv. aurantifolii or X.
campestris pv. citrumelo, confer ability to elicit hyperplasia (cell divisions
or cankers) on all citrus species in the normal host range of the recipient
strain.


|

|

|
X. campestris pv. aurantifolii |
| Host |
Common Name |
X. citri |
Pathotype B |
Pathotype C |

|

|

|

|

|
| C. sinensis |
Sweet Orange |
++ |
+ |
HR |
| C. paradisi |
Grapefruit |
++ |
+ |
HR |
| C. limon |
Lemon |
+ |
++ |
HR |
| C. aurantifolii |
Mexican Lime |
++ |
++C |
++ |

|

|

|

|

|

+ indicates virulence; ++ indicates strong virulence; HR indicates a hypersensitive response;
C indicates a distinctive chlorotic canker phenotype.

Pathotype B of X. campestris pv. aurantifolii causes "false" citrus canker or
cancrosis B, while Pathotype C causes Mexican lime cancrosis or cancrosis C.

VII. Pathogenicity

VIII. Storage of Organism

IX. Reported Host Range

X. Geographical Range and Spread

XI. Suggested Taxonomic Keys

XII. References

For general inquiries, please send e-mail to
info@ipgenetics.com. For web site errors or content issues, please e-mail
webmaster@ipgenetics.com.

 Copyright © October 2001 Integrated Plant Genetics, Inc. -- All Rights Reserved Copyright © October 2001 Integrated Plant Genetics, Inc. -- All Rights Reserved
|
|

|
|





